In our previous blog post OAK Cameras for NDVI Perception we explored NDVI approaches and how to calculate it using a multispectral camera.
Today, we are elevating (pun intended) the NDVI perception to the next level by using a drone with a multispectral camera and using SAM2 model for field segmentation and health comparison.
First: The hardware
We used the OAK-D-SR's PCBA and changed one CCM (Compact Camera Modules), so one sensor perceived the visible band (380-750nm) while the other perceived the NIR band (>750nm).
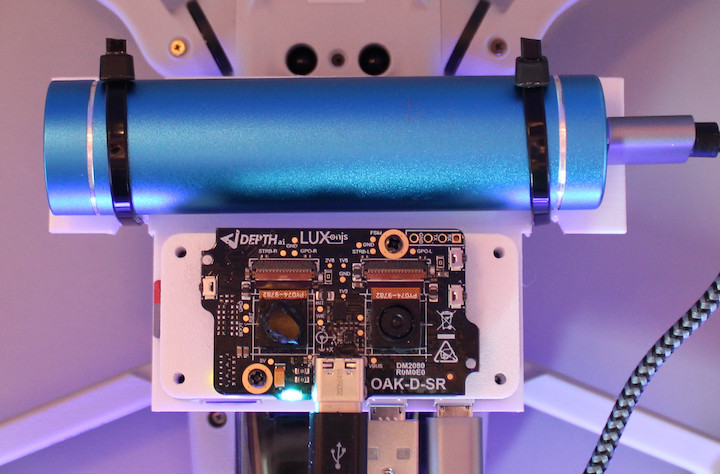
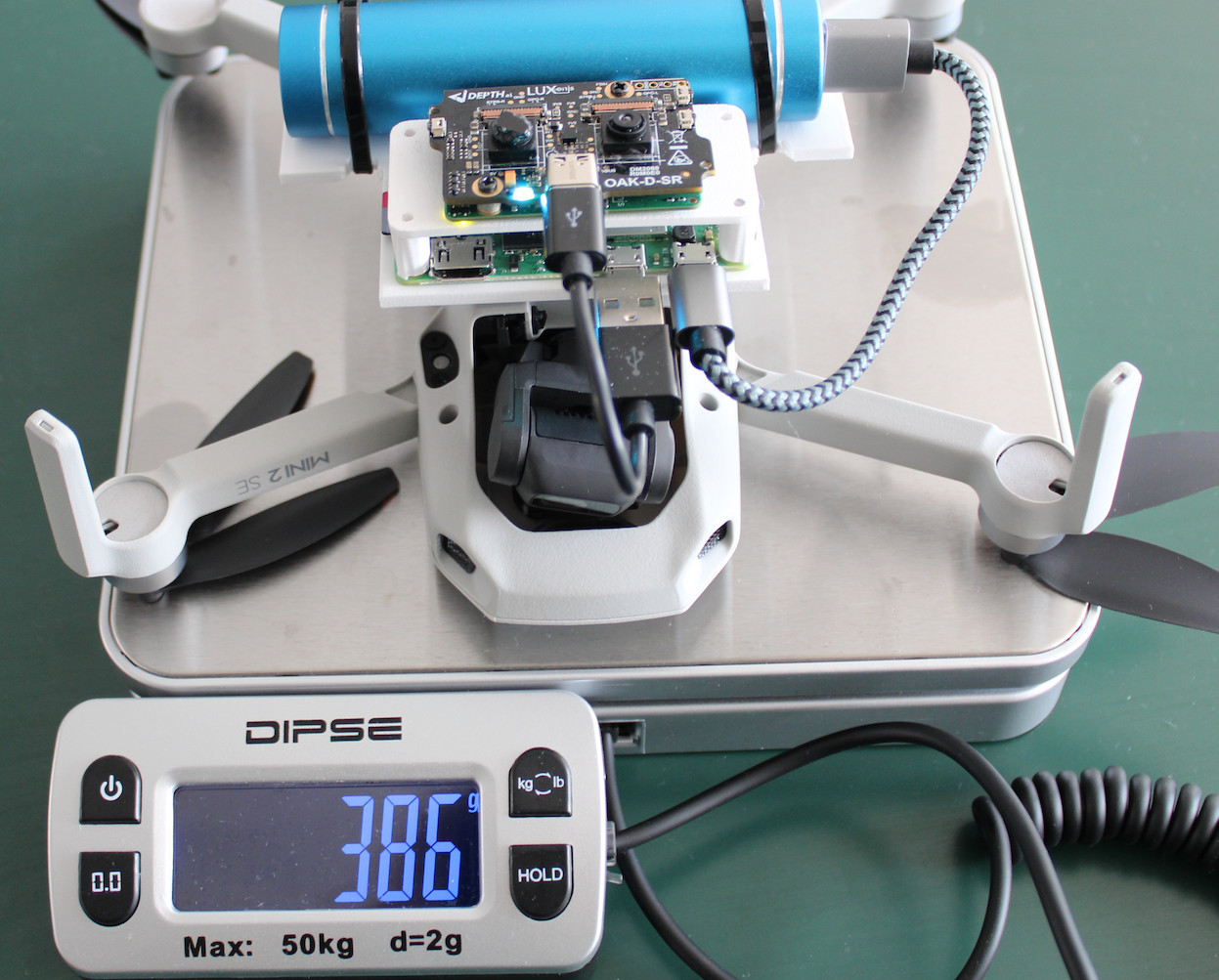
OAK-D-SR was connected to the RPi Zero 2W, which connected to the OAK-D-SR and saved both frames every second. Both of these devices were powered by a powerbank, and together with the DJI Mini 2 SE drone (249g), everything weighted 386g.
SAM2 segmentation
After recording left/right frames, I used SAM2 Demo app, uploaded the color video, and selected (only 3 at a time) fields we're interested in. After selecting the fields and running the model, you can check the segmentation results in the Networking tab, under "propagate_in_video" request. I saved these results in a file, and later decoded and visualized them.
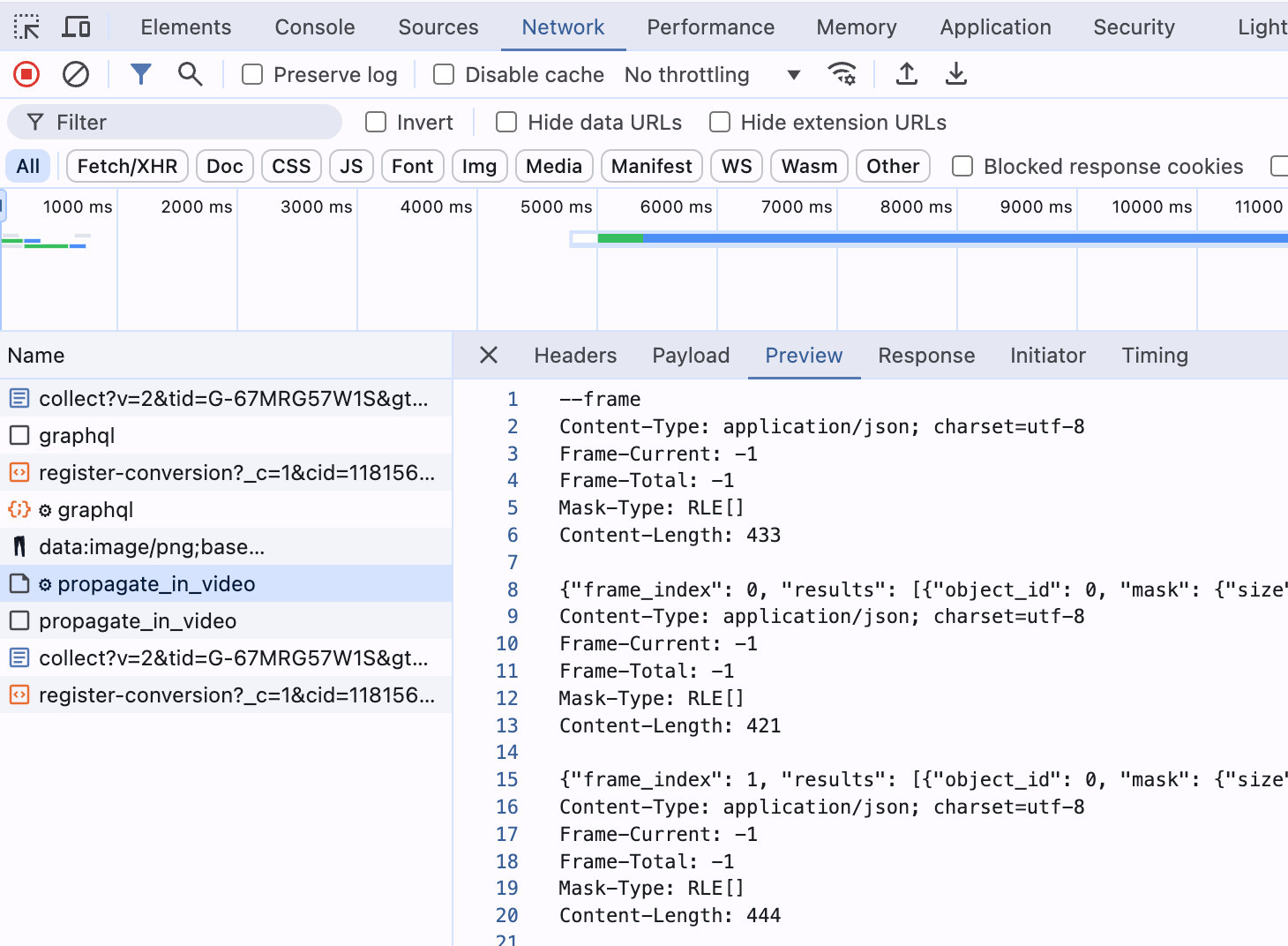
SAM2 results are in RLE encoded, so they need to be decoded to get the mask. You can use pycocotools to decode the mask.
from pycocotools import mask as mask_utils
mask = mask_utils.decode(annotation["segmentation"])
NDVI comparison
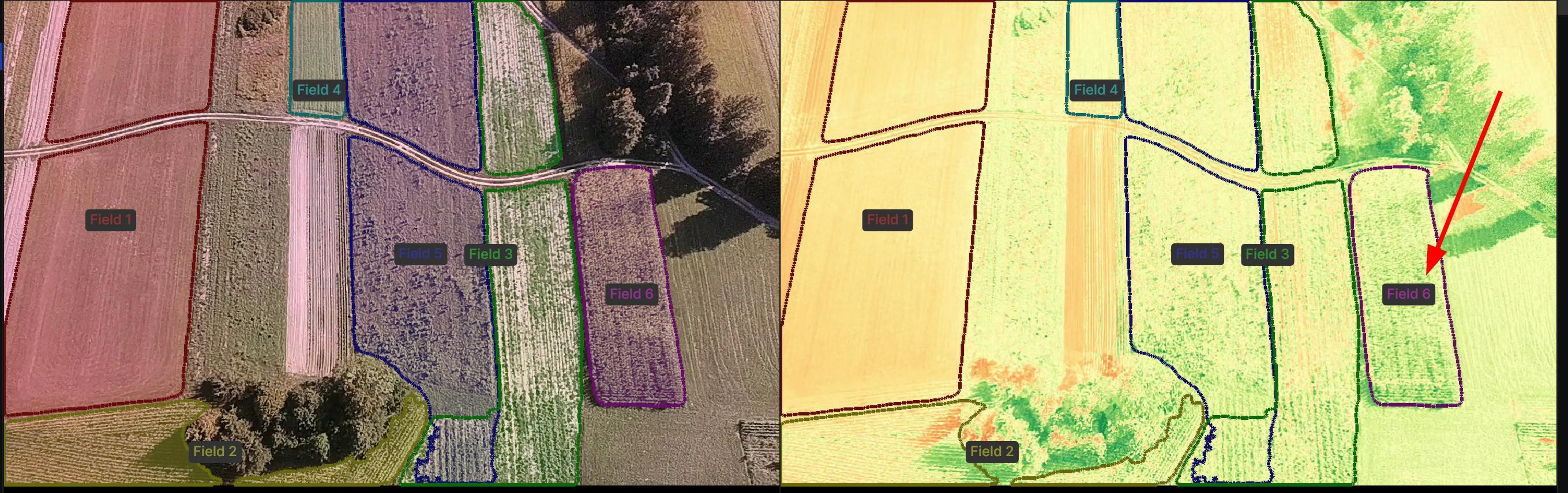
On the bottom of the demo you can see the NDVI comparison between the fields. Because NDVI is relative (not absolute), we can only use it to compare the health of the fields.
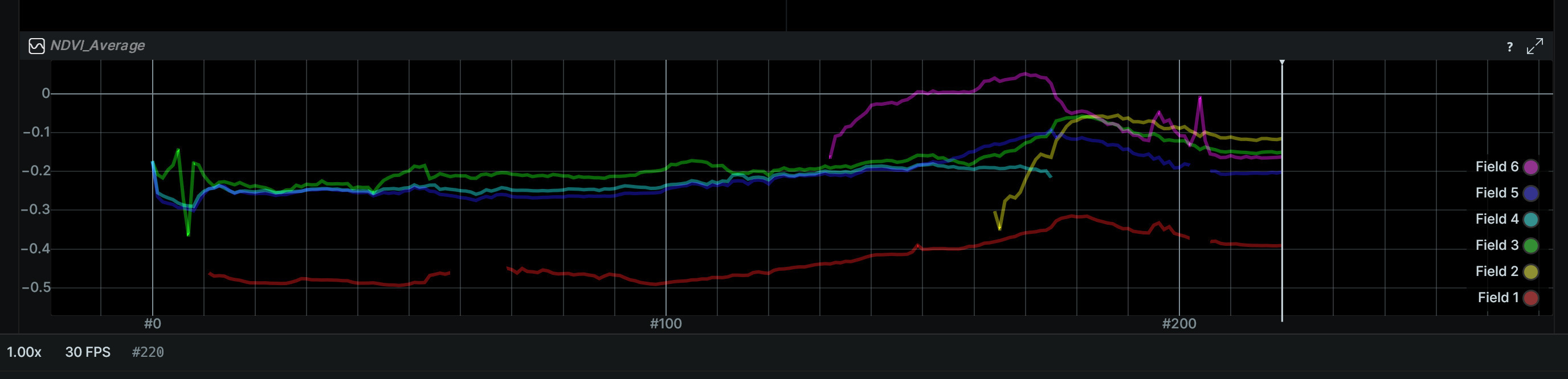
Field 6 has the highest NDVI value, which is also evident from the colorized NDVI image - it's more green than the other fields.
Visualization & Code
We're using Rerun for the whole visualization, and OpenCV for image processing and contour calculation (for nicer visualization).
Below is the main logic behind the demo. Full code can be found here, and full demo here (includes SAM results and videos).
# Run & initialize ReRun viewer
rr.init('NDVI /w SAM2', spawn=True)
# Prepare rerun visualization
annotationContext = [(0, "Background", (0, 0, 0, 0))]
for i, color in enumerate(colors):
annotationContext.append((i + 1, f"Field {i + 1}", color))
rr.log(f"NDVI_Average/Field{i+1}",
rr.SeriesLine(color=color, name=f"Field {i+1}")
)
rr.log("Color", rr.AnnotationContext(annotationContext), timeless=True)
t = 0
size = (800, 1280)
for frame_idx in range(len(sam_data[0])):
rr.set_time_sequence("step", t)
t += 1
frames = get_all_frames()
ndvi = calc_ndvi(frames['color'], frames['ir'])
segmentations = np.zeros(size)
for i, data in enumerate(get_sam_output(frame_idx)):
for result in data.get("results", []):
field_num = result['object_id']+i*3 # 3 segmentations per file
# Decode the RLE mask
mask = np.array(maskUtils.decode(result["mask"]), dtype=np.uint8)
# Set full_mask to num where mask
segmentations[mask == 1] = field_num + 1 # as 0 is Background
line_strips = get_contours(mask)
rr.log(
f"Color/Contours{field_num + 1}",
rr.LineStrips2D(line_strips, colors=colors[field_num])
)
rr.log(
f"NDVI/Color/Contours{field_num + 1}",
rr.LineStrips2D(line_strips, colors=colors[field_num]
)
mean_ndvi = np.mean(ndvi[mask == 1])
rr.log(f"NDVI_Average/Field{field_num + 1}", rr.Scalar(mean_ndvi))
rr.log("Color/Image", rr.Image(frames['color'][..., ::-1]))
rr.log("NDVI/Color", rr.Image(frames['ndvi_colorized'][..., ::-1]))
rr.log("Color/Mask", rr.SegmentationImage(segmentations))
Potential improvements
An important thing to note is that NDVI is calculated per image view, not globally. This is why the field's median NDVI changes instead of being a constant number. To improve this, we could use the whole video (eg. do image stitching) and calculate the NDVI for the entire area.
Conclusion
We've shown how to use a drone with a multispectral camera to capture NDVI images and use the SAM2 model for field segmentation and health comparison. This approach can be used for various agriculture tasks, such as monitoring crop health, detecting diseases, and more.
If you have any comments or suggestions, let me know in the comments!🙂